OHNOLOGS
A Repository of Genes Retained from Whole Genome Duplications in the Vertebrate Genomes
OHNOLOGS version 2.0 is now live with more genomes and more genome duplications!
OHNOLOGS is the database for the genes retained from Whole Genome Duplication in 27 vertebrate genomes. Ohnologs are gene duplicates originating from whole genome duplication (WGD). The name ohnolog has been coined after Susumu Ohno (1928-2000), who first proposed the two rounds of WGDs in the genome of vertebrate ancestor. Here you can download and explore ohnolog pairs and families in several vertebrate genomes computed with pre-defined or user-defined confidence levels (q-scores).
These ohnolog pairs and families have been constructed using a quantitative multiple-genome comparative approach, we had previously developed (Singh et al., PLOS Comp. Biol. 2015). The new revised version includes 27 vertebrates with ohnologs from 2R-WGD and 4 fish with ohnologs from 3R-WGD.
Species in OHNOLOGS v2 for the two rounds of Whole Genome Duplications in the ancestor of all verterates (2R WGD)
- Anole lizard (Anolis carolinensis)
|
 |
|
- Cat (Felis catus)
|
 |
|
- Chicken (Gallus gallus)
|
 |
|
- Chimpanzee (Pan troglodytes)
|
 |
- Cow (Bos taurus)
|
 |
|
- Dog (Canis familiaris)
|
 |
|
- Gorilla (Gorilla gorilla)
|
 |
|
- Horse (Equus caballus)
|
 |
- Human (Homo sapiens)
|
 |
|
- Macaque (Macaca mulatta)
|
 |
|
- Marmoset (Callithrix jacchus)
|
 |
|
- Medaka (Oryzias latipes)
|
 |
- Mouse (Mus musculus)
|
 |
|
- Olive baboon (Papio anubis)
|
 |
|
- Opossum (Monodelphis domestica)
|
 |
|
- Orangutan (Pongo abelii)
|
 |
- Pig (Sus scrofa)
|
 |
|
- Pufferfish (Tetraodon nigroviridis)
|
 |
|
- Rabbit (Oryctolagus cuniculus)
|
 |
|
- Rat (Rattus norvegicus)
|
 |
- Sheep Sheep (Ovis aries)
|
 |
|
- Spotted gar (Lepisosteus oculatus)
|
 |
|
- Stickleback (Gasterosteus aculeatus)
|
 |
|
- Turkey (Meleagris gallopavo)
|
 |
- Vervet-AGM (Chlorocebus sabaeus)
|
 |
|
- Zebra Finch (Taeniopygia guttata)
|
 |
|
- Zebrafish (Danio rerio)
|
 |
|
Species in OHNOLOGS v2 for the third round of Whole Genome Duplications in the ancestor of teleost fish (3R WGD)
- Medaka (Oryzias latipes)
|
 |
|
- Pufferfish (Tetraodon nigroviridis)
|
 |
|
- Stickleback (Gasterosteus aculeatus)
|
 |
|
- Zebrafish (Danio rerio)
|
 |
Why study ohnologs?
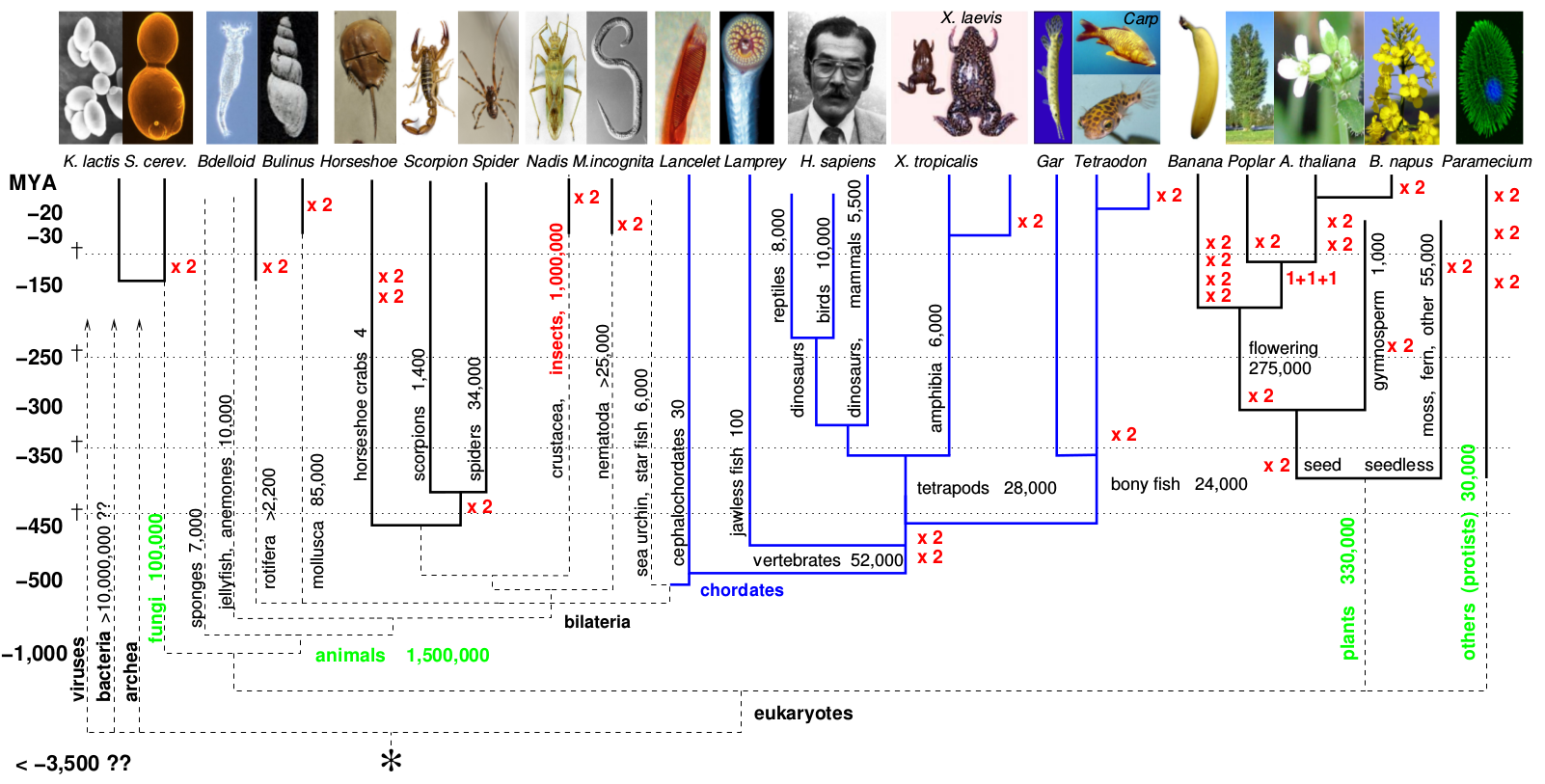
In many organisms descended from polyploid ancestors (shown in the figure below), ohnologs are associated with unique evolutionary innovations. For example, ohnologs are known to be associated with signaling pathways and developmental genes in vertebrates and most likely facilitated increased genomic, morphological and developmental complexity of vertebrates. In addition, ohnologs have been shown to present an enhanced susceptibility to deleterious mutations and are frequently associated with cancer and other genetic diseases. Therefore, these evolutionary observations also hold predictive power to identify and prioritize disease gene candidates and driver mutations in the NGS studies to sequence cancer genomes.
WGD on the Tree of Life
References
- Singh PP & Isambert H. (2019) OHNOLOGS v2: A comprehensive resource for the genes retained from whole genome duplication in vertebrates. Nucleic Acids Research. Full text
- Singh PP, Arora J & Isambert H. (2015) Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLOS Computational Biology. 11(7):e1004394. Full text
- Singh PP, Affeldt S, Cascone I, Selimoglu R, Camonis J & Isambert H. (2012) On the expansion of "dangerous" gene repertoires by whole-genome duplications in early vertebrates. Cell Reports. 2(5):1387-98. Full text
- Singh PP, Affeldt S, Malaguti G & Isambert H. (2014) Human Dominant Disease Genes are Enriched in Paralogs Originating from Whole Genome Duplication. PLOS Computational Biology. 10(7):e1003754. Full text
- Malaguti G, Singh PP & Isambert H. (2014) On the retention of gene duplicates prone to dominant deleterious mutations. Theoretical Population Biology. 93:38–51. Full text
 |
|
 |