OHNOLOGS
A Repository of Genes Retained from Whole Genome Duplications in the Vertebrate GenomesWhat's new in OHNOLOGS version 2.0?
- We have added 27 new vertebrate genomes with the two rounds of ancestral verterate Whole Genome Duplications (2R-WGD).
- There are 4 teleost fish genomes with the third round of ancestral teleost specific Whole Genome Duplication (3R-WGD).
- In addition to protein coding genes, OHNOLOGS v2 also includes non-coding RNA ohnologs (micro-RNA, sn-RNA, sno-RNA etc.)
- We have implemented a weighted confidence q-score taking into account the phylogenetically biased sampling of included vertebrate and teleost fish species (Singh and Isambert, Nucleic Acids Research, 2019).
- The code and more details for OHNOLOGS v2.0 can be found on the associated GitHub repository.
Algorithm
We identify ohnologs using a content-based macro-synteny comparison of a vertebrate genome with itself and with multiple outgroup genomes. An overall flowchart of our approach has been detailed below:
Q-score
To rule out spurious synteny, we developed a quantitative approach to assess the statistical confidence of each ohnolog pair (Singh et al. PLoS Comput Biol 2015). This quantitative approach and corresponding "q-score", ranging from 0 to 1, estimates the probability that each ohnolog pair is simply identified by chance. Hence, lower q-scores imply more statistically significant ohnolog pairs.
We filter and combine all the initial ohnolog candidates following the computational flowchart above and generate three sets of ohnolog pairs/families with varying level of statistical significance:
Understanding Ohnolog Families
A gene search leads to Ohnolog Family Page where the ohnolog families constructed by our approach are displayed. Ohnolog partners for the families are displayed in different columns. A typical ohnolog family is depicted below e.g. for the human NRAS gene

|
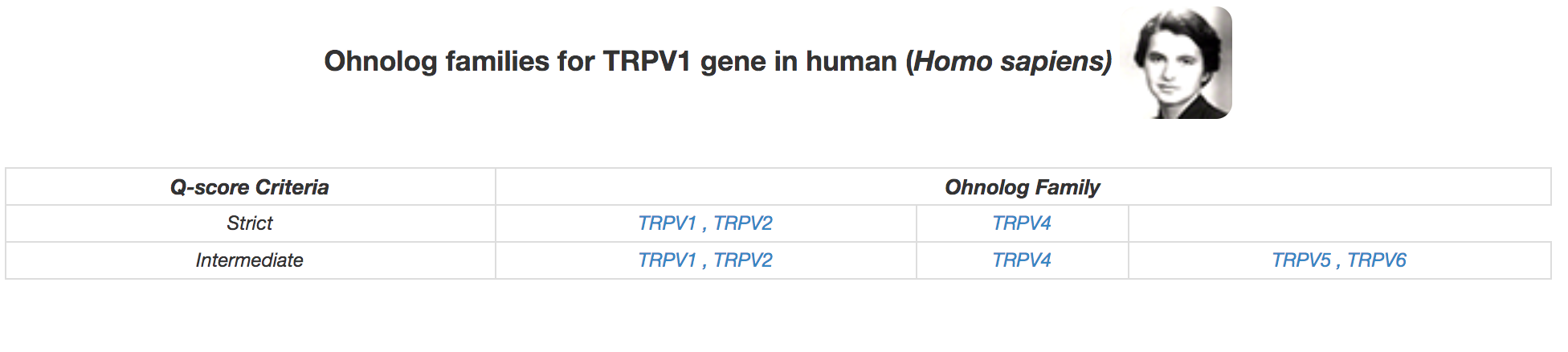
For any gene search, ohnolog families for the most stringent criteria are displayed. If relaxing the q-score criteria leads to a change in ohnolog family, we show them for multiple criteria. In the above example NRAS family is identified using the intermediate criteria. This family remains unchanged upon relaxing the q-score. However, in the example below, relaxing the q-score criteria for the human TRPV1 family gets and additional ohnologs "TRPV5, TRPV6" for the intermediate q-score criteria. There is no change in the family with the relaxed q-score and hence it has not been shown here.
|
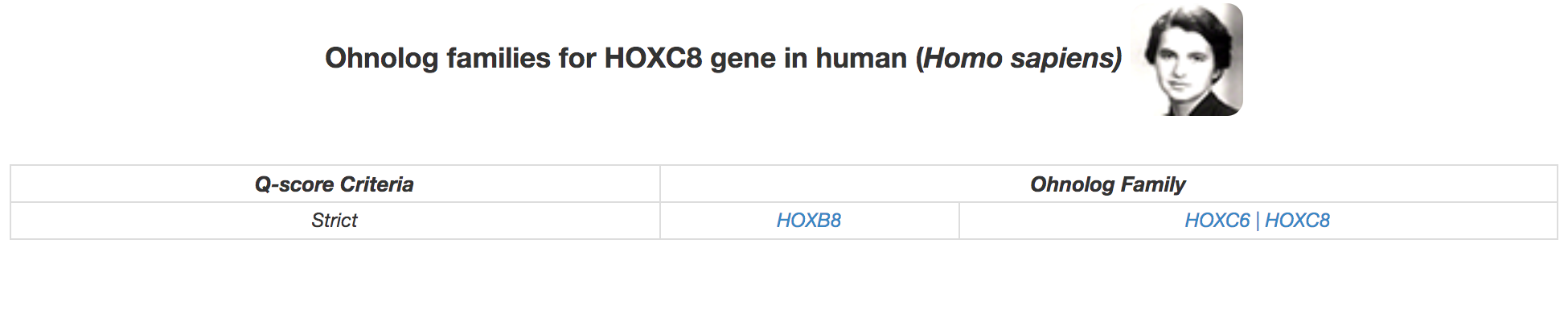
Genes within the same cell are small scale duplicates (SSDs) e.g. TRPV1, TRPV2 and TRPV5, TRPV6 in the example above. We use two different separators for SSDs: a comma (,) to distinguish if it is a recent SSD (after the WGD event), and a pipe (|) for an ancient SSD (before or around the same time as the WGD event). Hence, both TRPV1, TRPV2, and TRPV5, TRPV6 have been duplicated by a recent SSD. while HOXC6 and HOXC8 in the example below have been duplicated by an SSD older than 2R-WGD.
|
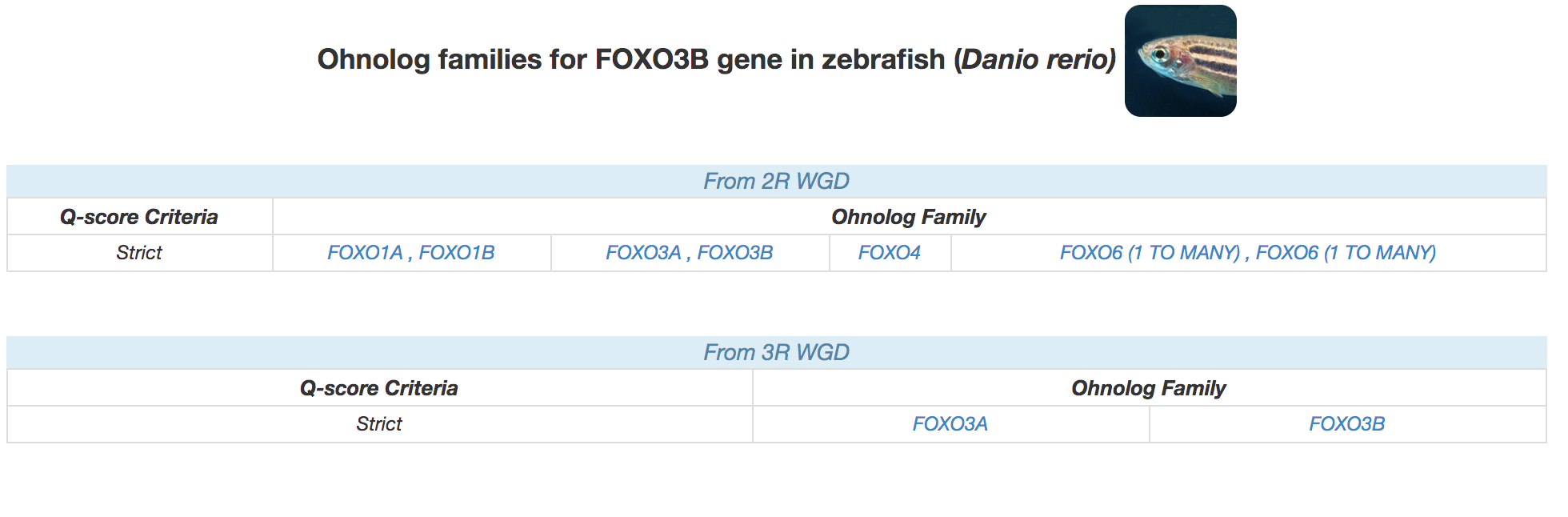
For the teleost fish species with an additional 3R-WGD, we display ohnologs from both 2R and 3R WGD like the example below for zebrafish FOXO3B family.
|
Generate Ohnolog Families
If you wish to generate custom ohnolog families using a different criteria, you can use generate functionality. For generating families, we start with all possible ohnolog pairs i.e. with any q-score and filter them based on user specifications. Then we run depth-first search to generate custom families and merge old and recent SSDs. These families, can be downloaded from the result page.
More details on the server and methods can be found in the published articles. We highly appreciate your feedback to improve the server. Please feel free to let us know if you find any bugs or if you have suggestions.
Download ohnolog pairs and families from older version 1
Click here if you wish to download the pairs and families from the older OHNOLOGS v1 (Singh et al. PLoS Comput Biol 2015).
 |
 |